Exploring a variant-adapted SARS-CoV-2 recombinant protein vaccine medrxivpreprint sanofi SARSCoV2 COVID19 Vaccine RecomibinantVaccine
By Bhavana KunkalikarDec 8 2022Reviewed by Aimee Molineux In a recent study posted to the medRxiv* preprint server, researchers assessed the immunogenicity of a variant-adapted severe acute respiratory syndrome coronavirus 2 recombinant protein vaccine boosted with an AS03 adjuvant.
In a significant change to the Phase 2 primary vaccine series trial on 10 June 2021, cohorts 1 and 2 were included for phase 3 assessment of the immunogenicity and safety of monovalent B.1.351, monovalent D614, and bivalent D614+B.1.351 CoV-2 preS dTM-AS03 booster preparations.
A lentivirus-based pseudovirus neutralization encoded the full-length spike protein of the D614G, Beta, or Omicron variants before neutralizing antibody responses to SARS-CoV-2 were quantified. The primary outcomes of the study involve individual serum PsVN titers against D614G at D1 and D15 for monovalent D614 vaccinees, against Beta for monovalent B.1.351 vaccinees, against B.1.351 and D614G for bivalent D614+B.1.
Belgique Dernières Nouvelles, Belgique Actualités
Similar News:Vous pouvez également lire des articles d'actualité similaires à celui-ci que nous avons collectés auprès d'autres sources d'information.
 Regression of Lung Cancer in Mice by Intranasal Administration of SARS-CoV-2 Spike S1This study underlines the importance of SARS-CoV-2 spike S1 in prompting death in cultured non-small cell lung cancer (NSCLC) cells and in vivo in lung tumors in mice. Interestingly, we found that recombinant spike S1 treatment at very low doses led to death of human A549 NSCLC cells. On the other hand, boiled recombinant SARS-CoV-2 spike S1 remained unable to induce death, suggesting that the induction of cell death in A549 cells was due to native SARS-CoV-2 spike S1 protein. SARS-CoV-2 spike S1-induced A549 cell death was also inhibited by neutralizing antibodies against spike S1 and ACE2. Moreover, our newly designed wild type ACE2-interacting domain of SARS-CoV-2 (wtAIDS), but not mAIDS, peptide also attenuated SARS-CoV-2 spike S1-induced cell death, suggesting that SARS-CoV-2 spike S1-induced death in A549 NSCLC cells depends on its interaction with ACE2 receptor. Similarly, recombinant spike S1 treatment also led to death of human H1299 and H358 NSCLC cells. Finally, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) intoxication led to the formation tumors in lungs of A/J mice and alternate day intranasal treatment with low dose of recombinant SARS-CoV-2 spike S1 from 22-weeks of NNK insult (late stage) induced apoptosis and tumor regression in the lungs. These studies indicate that SARS-CoV-2 spike S1 may have implications for lung cancer treatment.
Regression of Lung Cancer in Mice by Intranasal Administration of SARS-CoV-2 Spike S1This study underlines the importance of SARS-CoV-2 spike S1 in prompting death in cultured non-small cell lung cancer (NSCLC) cells and in vivo in lung tumors in mice. Interestingly, we found that recombinant spike S1 treatment at very low doses led to death of human A549 NSCLC cells. On the other hand, boiled recombinant SARS-CoV-2 spike S1 remained unable to induce death, suggesting that the induction of cell death in A549 cells was due to native SARS-CoV-2 spike S1 protein. SARS-CoV-2 spike S1-induced A549 cell death was also inhibited by neutralizing antibodies against spike S1 and ACE2. Moreover, our newly designed wild type ACE2-interacting domain of SARS-CoV-2 (wtAIDS), but not mAIDS, peptide also attenuated SARS-CoV-2 spike S1-induced cell death, suggesting that SARS-CoV-2 spike S1-induced death in A549 NSCLC cells depends on its interaction with ACE2 receptor. Similarly, recombinant spike S1 treatment also led to death of human H1299 and H358 NSCLC cells. Finally, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) intoxication led to the formation tumors in lungs of A/J mice and alternate day intranasal treatment with low dose of recombinant SARS-CoV-2 spike S1 from 22-weeks of NNK insult (late stage) induced apoptosis and tumor regression in the lungs. These studies indicate that SARS-CoV-2 spike S1 may have implications for lung cancer treatment.
Lire la suite »
 Predictive model for long COVID in children 3 months after a SARS-CoV-2 PCR test - BMC MedicineBackground To update and internally validate a model to predict children and young people (CYP) most likely to experience long COVID (i.e. at least one impairing symptom) 3 months after SARS-CoV-2 PCR testing and to determine whether the impact of predictors differed by SARS-CoV-2 status. Methods Data from a nationally matched cohort of SARS-CoV-2 test-positive and test-negative CYP aged 11–17 years was used. The main outcome measure, long COVID, was defined as one or more impairing symptoms 3 months after PCR testing. Potential pre-specified predictors included SARS-CoV-2 status, sex, age, ethnicity, deprivation, quality of life/functioning (five EQ-5D-Y items), physical and mental health and loneliness (prior to testing) and number of symptoms at testing. The model was developed using logistic regression; performance was assessed using calibration and discrimination measures; internal validation was performed via bootstrapping and the final model was adjusted for overfitting. Results A total of 7139 (3246 test-positives, 3893 test-negatives) completing a questionnaire 3 months post-test were included. 25.2% (817/3246) of SARS-CoV-2 PCR-positives and 18.5% (719/3893) of SARS-CoV-2 PCR-negatives had one or more impairing symptoms 3 months post-test. The final model contained SARS-CoV-2 status, number of symptoms at testing, sex, age, ethnicity, physical and mental health, loneliness and four EQ-5D-Y items before testing. Internal validation showed minimal overfitting with excellent calibration and discrimination measures (optimism-adjusted calibration slope: 0.96575; C-statistic: 0.83130). Conclusions We updated a risk prediction equation to identify those most at risk of long COVID 3 months after a SARS-CoV-2 PCR test which could serve as a useful triage and management tool for CYP during the ongoing pandemic. External validation is required before large-scale implementation.
Predictive model for long COVID in children 3 months after a SARS-CoV-2 PCR test - BMC MedicineBackground To update and internally validate a model to predict children and young people (CYP) most likely to experience long COVID (i.e. at least one impairing symptom) 3 months after SARS-CoV-2 PCR testing and to determine whether the impact of predictors differed by SARS-CoV-2 status. Methods Data from a nationally matched cohort of SARS-CoV-2 test-positive and test-negative CYP aged 11–17 years was used. The main outcome measure, long COVID, was defined as one or more impairing symptoms 3 months after PCR testing. Potential pre-specified predictors included SARS-CoV-2 status, sex, age, ethnicity, deprivation, quality of life/functioning (five EQ-5D-Y items), physical and mental health and loneliness (prior to testing) and number of symptoms at testing. The model was developed using logistic regression; performance was assessed using calibration and discrimination measures; internal validation was performed via bootstrapping and the final model was adjusted for overfitting. Results A total of 7139 (3246 test-positives, 3893 test-negatives) completing a questionnaire 3 months post-test were included. 25.2% (817/3246) of SARS-CoV-2 PCR-positives and 18.5% (719/3893) of SARS-CoV-2 PCR-negatives had one or more impairing symptoms 3 months post-test. The final model contained SARS-CoV-2 status, number of symptoms at testing, sex, age, ethnicity, physical and mental health, loneliness and four EQ-5D-Y items before testing. Internal validation showed minimal overfitting with excellent calibration and discrimination measures (optimism-adjusted calibration slope: 0.96575; C-statistic: 0.83130). Conclusions We updated a risk prediction equation to identify those most at risk of long COVID 3 months after a SARS-CoV-2 PCR test which could serve as a useful triage and management tool for CYP during the ongoing pandemic. External validation is required before large-scale implementation.
Lire la suite »
 Researchers explore the correlation between the dynamic shedding pattern of SARS-CoV-2 and viral loadResearchers attempted to establish an association between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load, indicating its ribonucleic acid (RNA) levels, and the presence of infectious virions.
Researchers explore the correlation between the dynamic shedding pattern of SARS-CoV-2 and viral loadResearchers attempted to establish an association between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load, indicating its ribonucleic acid (RNA) levels, and the presence of infectious virions.
Lire la suite »
 Study explores the risk of new-onset diabetes mellitus following SARS-CoV-2 infectionsResearchers evaluated individuals who had SARS-CoV-2 infections and were diagnosed with diabetes mellitus within six months of the onset of COVID-19 to understand the temporal relationship between SARS-CoV-2 infections and diabetes mellitus.
Study explores the risk of new-onset diabetes mellitus following SARS-CoV-2 infectionsResearchers evaluated individuals who had SARS-CoV-2 infections and were diagnosed with diabetes mellitus within six months of the onset of COVID-19 to understand the temporal relationship between SARS-CoV-2 infections and diabetes mellitus.
Lire la suite »
 Machine learning analysis suggests that there are four sub-phenotypes of long COVIDMachine learning analysis suggests that there are four sub-phenotypes of long COVID NatureMedicine WeillCornell longCOVID coronavirus covid machinelearning data healthcaredata phenotype
Machine learning analysis suggests that there are four sub-phenotypes of long COVIDMachine learning analysis suggests that there are four sub-phenotypes of long COVID NatureMedicine WeillCornell longCOVID coronavirus covid machinelearning data healthcaredata phenotype
Lire la suite »
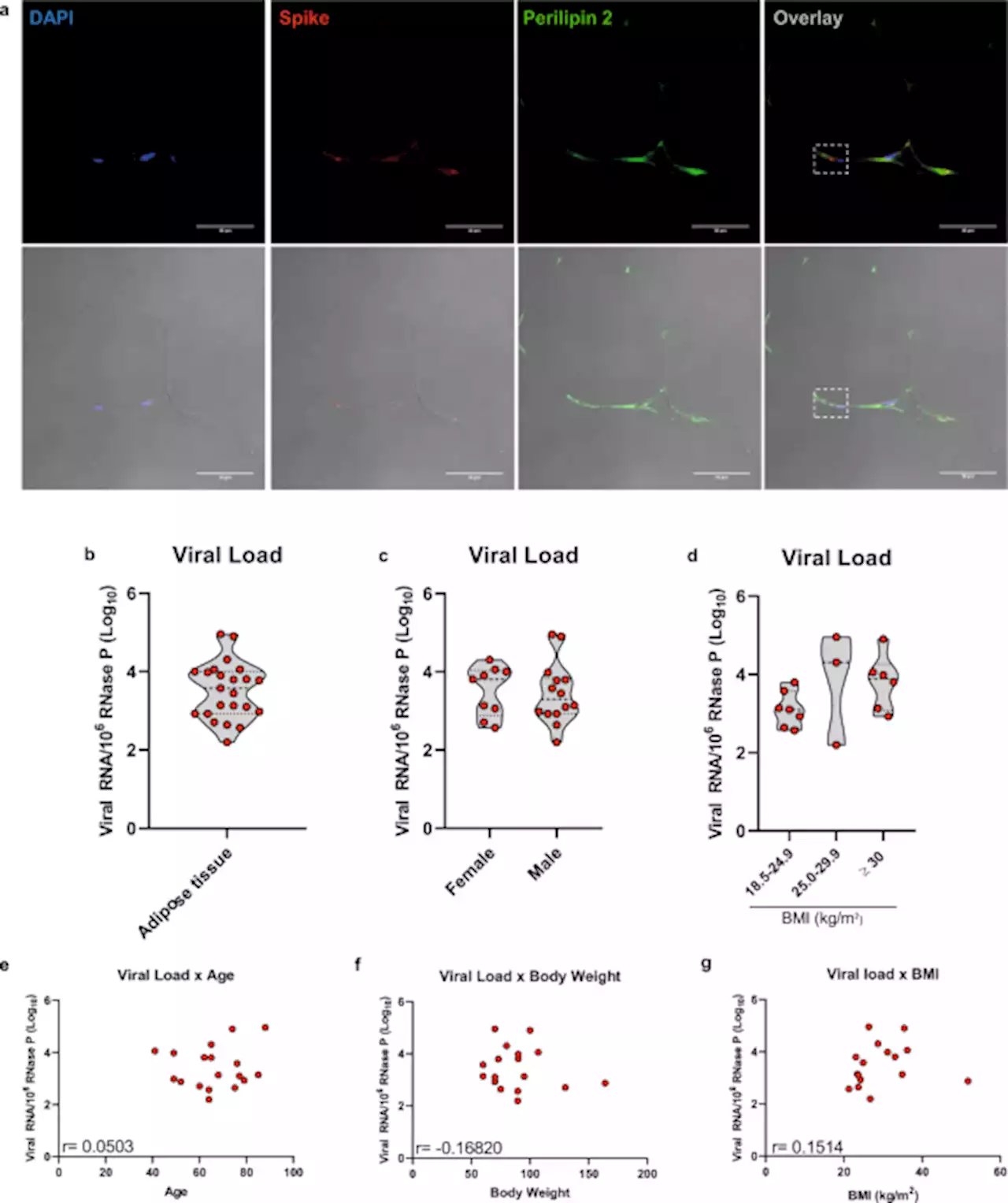 SARS-CoV-2 infects adipose tissue in a fat depot- and viral lineage-dependent manner - Nature CommunicationsVisceral adiposity is a risk factor for severe COVID-19, and infection of adipose tissue by SARS-CoV-2 has been reported. Here the authors confirm that human adipose tissue is a possible site for SARS-CoV-2 infection, but the degree of adipose tissue infection and the way adipocytes respond to the virus depend on the adipose tissue depot and the viral strain.
SARS-CoV-2 infects adipose tissue in a fat depot- and viral lineage-dependent manner - Nature CommunicationsVisceral adiposity is a risk factor for severe COVID-19, and infection of adipose tissue by SARS-CoV-2 has been reported. Here the authors confirm that human adipose tissue is a possible site for SARS-CoV-2 infection, but the degree of adipose tissue infection and the way adipocytes respond to the virus depend on the adipose tissue depot and the viral strain.
Lire la suite »
